Juniper Publishers - Blood-Drop Liquid Biopsy for Monitoring Mutation Load and Therapeutic Responses
Cancer Therapy & Oncology International Journal
Abstract
Tumor heterogeneity, especially intra-tumor
heterogeneity, is a crucial factor underlying difficulties in cancer
treatment and failure of a number of current therapeutic modalities,
even of molecularly targeted therapies. The conventional follow-up care
is based on regular observation of protein markers in combination with
computed tomography (CT)/positron emission tomography (PET) imaging to
monitor stable disease or tumor progression. More recently, the
implementation of a noninvasive “liquid biopsy” has been destined to
capture tumor-specific genetic clonal evolution throughout the course of
cancer therapy. This case report focuses on circulating cell-free tumor
DNA (ctDNA) in the bloodstream as a versatile biomarker, enabling
accurate monitoring of tumor burden and treatment response. Our
observations showed that [1] mutation load measured by blood-drop ctDNA
sequencing correlated well with PET/CT, CA biomarkers and clinical
outcome in response to therapy; [2] blood-drop ctDNA testing could
detect new tumor mass earlier than PET/CT; [3] blood-drop liquid biopsy
possessed longitudinal and real-time monitoring capability; [4]
blood-drop liquid biopsy could be a surrogate for accurate, affordable
and accessible personalized testing.
Keywords: Blood-Drop; Liquid Biopsy; Cell-free tumor DNA; Cancer Therapy
Abbreviations: CT: Computed Tomography; PET: Positron Emission Tomography; ctDNA: Cell-Free Tumor DNA
Tumor heterogeneity, especially intra-tumor heterogeneity, is a crucial factor underlying difficulties in cancer treatment and failure of a number of current therapeutic modalities, even of molecularly targeted therapies. The conventional follow-up care is based on regular observation of protein markers in combination with computed tomography (CT)/positron emission tomography (PET) imaging to monitor stable disease or tumor progression. More recently, the implementation of a noninvasive “liquid biopsy” has been destined to capture tumor-specific genetic clonal evolution throughout the course of cancer therapy. This case report focuses on circulating cell-free tumor DNA (ctDNA) in the bloodstream as a versatile biomarker, enabling accurate monitoring of tumor burden and treatment response. Our observations showed that [1] mutation load measured by blood-drop ctDNA sequencing correlated well with PET/CT, CA biomarkers and clinical outcome in response to therapy; [2] blood-drop ctDNA testing could detect new tumor mass earlier than PET/CT; [3] blood-drop liquid biopsy possessed longitudinal and real-time monitoring capability; [4] blood-drop liquid biopsy could be a surrogate for accurate, affordable and accessible personalized testing.
Keywords: Blood-Drop; Liquid Biopsy; Cell-free tumor DNA; Cancer Therapy
Abbreviations: CT: Computed Tomography; PET: Positron Emission Tomography; ctDNA: Cell-Free Tumor DNA
Introduction
Tumor genome sequencing to inform treatment decisions
is the current standard-of-care to personalized cancer management
[1,2]. Tumor tissue biopsy, generally from the primary site, is used to
determine molecular alterations at a single time point on a single site,
before targeted treatment commences. These biopsies carry potential
risks for patients, they are painful, costly with high failure rate and
unpredictable turnaround time. Most importantly, given the complexities
of tumor dynamic heterogeneity, both within a tumor and between a
primary tumor and metastases, a tissue sample may not be a true
representation of tumor genomic evolution [3-5].
Therefore, inability to capture spatial and temporal
heterogeneity during tumor evolution results in the failure of cancer
systemic treatments, requiring the development of novel approaches to
better track tumor heterogeneity.
Circulating protein biomarkers are traditionally used
in cancer diagnosis and in the assessment of therapeutic responses,such
as carcinoma embryonic antigen (CEA), prostate-specific antigen (PSA),
cancer antigen (CA) 3-15, 9-19, 27-29, and CA-125. Unfortunately, the
specificity and reliability of these markers are not satisfactory, and
many malignancies even do not have released any reliable protein
biomarker [6,7].
The advantage of clinical PET/CT imaging for
monitoring tumor response replies upon its ability to interrogate both
anatomic and functional measures of treatment effects. Although the
resolution of this technology has been greatly improved over years, the
reality is even a tiny lesion (approx. 1 cm3) spotted on the image could represent billions of tumor cells.
Circulating cell-free tumor DNA (ctDNA) carries
comprehensive, inherently specific, and highly sensitive information,
and thus possesses distinctive advantage over conventional protein
biomarkers and imaging technology. Studies in melanoma [8], breast [9],
ovarian [10], and colon cancers [11] have validated the potential
applications of ctDNA to monitor tumor burden dynamically and precisely
during treatment process. A liquid biopsy based on ctDNA could capture
the entire heterogeneity of the disease longitudinally as well as in
real time
[12].
This will allow clinicians to ensure that the therapy they have
selected, based on a particular molecular target, remains relevant
and observe the emergence of any resistance [13]. Instead of
waiting for information from imaging scans “reactively”, doctors
could “proactively” identify at an earlier stage whether a treatment
is not working and to spare the patient with the unnecessary
toxicity of a drug that no longer provides any benefit. Likewise,
physicians could monitor if any new molecular targets emerging
and adjust treatment strategy accordingly. This global picture not
only can help for designing combination treatments to minimize
therapeutic resistance, but also help to provide patients with the
right treatment for the right target without delay.
Tumor genome sequencing to inform treatment decisions is the current standard-of-care to personalized cancer management [1,2]. Tumor tissue biopsy, generally from the primary site, is used to determine molecular alterations at a single time point on a single site, before targeted treatment commences. These biopsies carry potential risks for patients, they are painful, costly with high failure rate and unpredictable turnaround time. Most importantly, given the complexities of tumor dynamic heterogeneity, both within a tumor and between a primary tumor and metastases, a tissue sample may not be a true representation of tumor genomic evolution [3-5].
Therefore, inability to capture spatial and temporal heterogeneity during tumor evolution results in the failure of cancer systemic treatments, requiring the development of novel approaches to better track tumor heterogeneity.
Circulating protein biomarkers are traditionally used in cancer diagnosis and in the assessment of therapeutic responses,such as carcinoma embryonic antigen (CEA), prostate-specific antigen (PSA), cancer antigen (CA) 3-15, 9-19, 27-29, and CA-125. Unfortunately, the specificity and reliability of these markers are not satisfactory, and many malignancies even do not have released any reliable protein biomarker [6,7].
The advantage of clinical PET/CT imaging for monitoring tumor response replies upon its ability to interrogate both anatomic and functional measures of treatment effects. Although the resolution of this technology has been greatly improved over years, the reality is even a tiny lesion (approx. 1 cm3) spotted on the image could represent billions of tumor cells.
Circulating cell-free tumor DNA (ctDNA) carries comprehensive, inherently specific, and highly sensitive information, and thus possesses distinctive advantage over conventional protein biomarkers and imaging technology. Studies in melanoma [8], breast [9], ovarian [10], and colon cancers [11] have validated the potential applications of ctDNA to monitor tumor burden dynamically and precisely during treatment process. A liquid biopsy based on ctDNA could capture the entire heterogeneity of the disease longitudinally as well as in real time [12].
This will allow clinicians to ensure that the therapy they have selected, based on a particular molecular target, remains relevant and observe the emergence of any resistance [13]. Instead of waiting for information from imaging scans “reactively”, doctors could “proactively” identify at an earlier stage whether a treatment is not working and to spare the patient with the unnecessary toxicity of a drug that no longer provides any benefit. Likewise, physicians could monitor if any new molecular targets emerging and adjust treatment strategy accordingly. This global picture not only can help for designing combination treatments to minimize therapeutic resistance, but also help to provide patients with the right treatment for the right target without delay.
Materials and Methods
Serum CEA, CA 19-9, 15-3, 27-29, and CA-125 levels were
determined by quantitative immunoassay. Blood-drop cellfree
DNA was enriched from 20 uL of plasma using Circulogene
proprietary technology. DNA concentrations were measured by
Qubit fluorometer using dsDNA BR kit. Optimal concentration
of 1-10 ng was used to amplify 207 targeted loci using AmpliSeq
Cancer Hotspot Panel, version 2 (Life Technologies, Carlsbad, CA,
USA), targeted for 2855 hotspot mutations within the 50 cancer
driver genes, according to the supplier’s protocol. Subsequent semiconductor-based sequencing was performed on Ion Chef and
Ion Proton (Life Technologies, Carlsbad, CA, USA), maintaining the
number of reads as >200,000 per sample. Base calling and data
analysis was performed as described [14].
Serum CEA, CA 19-9, 15-3, 27-29, and CA-125 levels were determined by quantitative immunoassay. Blood-drop cellfree DNA was enriched from 20 uL of plasma using Circulogene proprietary technology. DNA concentrations were measured by Qubit fluorometer using dsDNA BR kit. Optimal concentration of 1-10 ng was used to amplify 207 targeted loci using AmpliSeq Cancer Hotspot Panel, version 2 (Life Technologies, Carlsbad, CA, USA), targeted for 2855 hotspot mutations within the 50 cancer driver genes, according to the supplier’s protocol. Subsequent semiconductor-based sequencing was performed on Ion Chef and Ion Proton (Life Technologies, Carlsbad, CA, USA), maintaining the number of reads as >200,000 per sample. Base calling and data analysis was performed as described [14].
Results and Discussion
We performed longitudinal monitoring of somatic alterations
in plasma ctDNA, i.e., mutation load, during therapy in 2 patients.
The dynamics of mutation load was plotted and compared with
CA markers and PET/CT imaging over the course of treatment.
Imaging scans were evaluated using the Response Evaluation
Criteria in Solid Tumors (RECIST; [15]).
Details of individual cases are provided below.
We performed longitudinal monitoring of somatic alterations in plasma ctDNA, i.e., mutation load, during therapy in 2 patients. The dynamics of mutation load was plotted and compared with CA markers and PET/CT imaging over the course of treatment. Imaging scans were evaluated using the Response Evaluation Criteria in Solid Tumors (RECIST; [15]).
Details of individual cases are provided below.
Patient #1
A 69-year-old non-smoker Chinese female patient with
diagnosis of lung cancer with metastasis. Tissue biopsy showed
sensitizing EGFR mutation, while ALK, ROS-1, BRAF were negative
on May 19, 2015. She was then placed on Tarceva 100 mg daily
and continued with Avastin once a month. After near 5-month
of targeted drug administration, her CEA, CA19-9, CA125 levels
all dropped significantly (Figure 1, left panel), and PET/CT scan
evaluation also showed stable disease that was maintained for
101 days with decreases in tumor size and activity (Figure 1, right
panel).
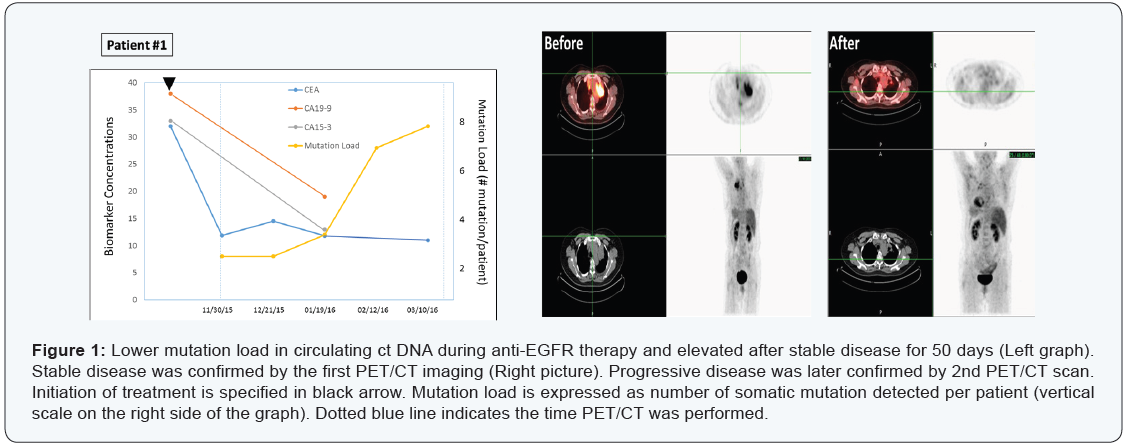
On the other hand, plasma ctDNA mutation dynamics detected
a new progression. Two somatic mutations was maintained for
about 50 days (in TP53 and PTEN genes), then gradually increased
to 3, 7 and 8 mutations by the end of March 2016, indicating a
progressive disease.
The mutation analysis on March 10th revealed 2 low-allelefrequency
sub clones of EGFR - E114K (4.2%) and E868G (2.4%),
implying a clonally evolution upon selection pressure by the drugs.
Most importantly, PET/CT scans on March 31, 2016 identified a
new tumor mass (about 1 cm3), confirming what ct DNA mutation
analysis has found earlier (Figure 1) upper panel.
Overall, this case demonstrated that the ctDNA “genetic
responses” were closely associated with radiologically stable
disease, with increases in the mutation load emerging ~2 months
earlier than radiological progression.
A 69-year-old non-smoker Chinese female patient with diagnosis of lung cancer with metastasis. Tissue biopsy showed sensitizing EGFR mutation, while ALK, ROS-1, BRAF were negative on May 19, 2015. She was then placed on Tarceva 100 mg daily and continued with Avastin once a month. After near 5-month of targeted drug administration, her CEA, CA19-9, CA125 levels all dropped significantly (Figure 1, left panel), and PET/CT scan evaluation also showed stable disease that was maintained for 101 days with decreases in tumor size and activity (Figure 1, right panel).

On the other hand, plasma ctDNA mutation dynamics detected a new progression. Two somatic mutations was maintained for about 50 days (in TP53 and PTEN genes), then gradually increased to 3, 7 and 8 mutations by the end of March 2016, indicating a progressive disease.
The mutation analysis on March 10th revealed 2 low-allelefrequency sub clones of EGFR - E114K (4.2%) and E868G (2.4%), implying a clonally evolution upon selection pressure by the drugs. Most importantly, PET/CT scans on March 31, 2016 identified a new tumor mass (about 1 cm3), confirming what ct DNA mutation analysis has found earlier (Figure 1) upper panel.
Overall, this case demonstrated that the ctDNA “genetic responses” were closely associated with radiologically stable disease, with increases in the mutation load emerging ~2 months earlier than radiological progression.
Patient #2
A 79-year-old Iranian female patient with diagnosis of
metastatic peri-pancreatic lymph node adenocarcinoma with
unknown primary. Immunohistochemistry on fine-needle biopsy
revealed CK7 positive, while CK20, TTF, S100 and CD45 all negative
on October 26, 2015. She was placed on XELOX initially. Later immunostaining also identified PD-L1 overexpression, therefore,
she was then treated with XELIRI, Avastin, and Opdivo. PET/CT
scans on February 11, 2016 showed significant decrease in tumor
size and activity with >90% response rate (Figure 2, right panel).
Results from mutation load by plasma ctDNA sequencing were in
agreement with those of imaging, CEA and CA markers (CA125,
CA27-29, CA19-9), indicating a stable disease maintained at least
for 41 days (Figure 2, left panel).
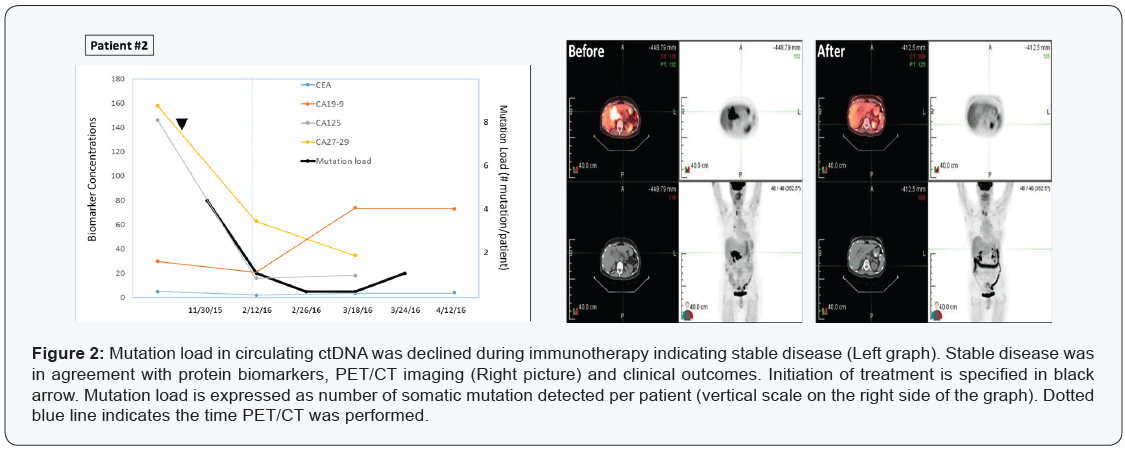
Four somatic mutations were detected initially (FLT3 Y572C
6.5%; TP53 E165G 5.2%; TP53 Y104C 4.1%; TP53 C137Y
4.0%), then declined to 1 and zero mutation during the course
of treatment. This case illustrated again the mutation analysis
by blood-drop liquid biopsy correlated strongly with clinical
outcomes in response to therapy.
The CEA and CA protein markers are not necessarily specific
to cancer cells and PET/CT scans are suffering from resolution
limitation. By contrast, cancer-associated somatic mutations are
specific to malignancies, and plasma DNA with these mutations is
indicative for the presence of malignancies. Our results on these
two cases support the notion that levels of the somatic mutations
detected from blood correlated well with current standard care
test results and clinical outcomes, i.e., stable disease or progressive
disease, and may provide the earliest indication of recurrence.
A 79-year-old Iranian female patient with diagnosis of metastatic peri-pancreatic lymph node adenocarcinoma with unknown primary. Immunohistochemistry on fine-needle biopsy revealed CK7 positive, while CK20, TTF, S100 and CD45 all negative on October 26, 2015. She was placed on XELOX initially. Later immunostaining also identified PD-L1 overexpression, therefore, she was then treated with XELIRI, Avastin, and Opdivo. PET/CT scans on February 11, 2016 showed significant decrease in tumor size and activity with >90% response rate (Figure 2, right panel).
Results from mutation load by plasma ctDNA sequencing were in agreement with those of imaging, CEA and CA markers (CA125, CA27-29, CA19-9), indicating a stable disease maintained at least for 41 days (Figure 2, left panel).

Four somatic mutations were detected initially (FLT3 Y572C 6.5%; TP53 E165G 5.2%; TP53 Y104C 4.1%; TP53 C137Y 4.0%), then declined to 1 and zero mutation during the course of treatment. This case illustrated again the mutation analysis by blood-drop liquid biopsy correlated strongly with clinical outcomes in response to therapy.
The CEA and CA protein markers are not necessarily specific to cancer cells and PET/CT scans are suffering from resolution limitation. By contrast, cancer-associated somatic mutations are specific to malignancies, and plasma DNA with these mutations is indicative for the presence of malignancies. Our results on these two cases support the notion that levels of the somatic mutations detected from blood correlated well with current standard care test results and clinical outcomes, i.e., stable disease or progressive disease, and may provide the earliest indication of recurrence.
Conclusion
We have demonstrated for the first time that blood-drop
ctDNA sequencing test can detect the presence of new tumor mass
earlier than current standard care imaging methods. The mutation
detection of ctDNA in drops of blood is a powerful monitoring tool
capable of providing accurate and earlier assessment of tumor
behavior, burden and patient responses following treatment.
To Know More
About Cancer
Therapy & Oncology International Journal Please click on:
https://juniperpublishers.com/ctoij/index.php
To Know More About Open
Access Journals Please click on:
https://juniperpublishers.com/index.php
We have demonstrated for the first time that blood-drop ctDNA sequencing test can detect the presence of new tumor mass earlier than current standard care imaging methods. The mutation detection of ctDNA in drops of blood is a powerful monitoring tool capable of providing accurate and earlier assessment of tumor behavior, burden and patient responses following treatment.
To Know More
About Cancer
Therapy & Oncology International Journal Please click on:
https://juniperpublishers.com/ctoij/index.php
To Know More About Open
Access Journals Please click on:
https://juniperpublishers.com/index.php



Comments
Post a Comment